Perspectives on the development of the inPROBE technology platform for protein expression detection
May 17, 2024
The innovative technological platform inPROBE® has the potential to be applied in various diagnostic fields. It utilizes a completely new approach to protein expression detection. This method enables in vivo diagnostics, meaning it can be performed directly within the patient's body without the need for tissue sampling. An objective numerical result is available almost immediately, within approximately 30 minutes. After summarizing the first phase of clinical trials, the company is now looking toward further applications of this groundbreaking technology.
An inherent feature of modern oncology is personalization, which manifests in the transition from a "one-size-fits-all" approach—using the same chemotherapy regimens for all patients with a specific type of cancer—to therapy tailored to the individual patient's situation. A crucial element of personalization is the development of molecularly targeted therapies, also known as targeted therapies, which act on specific molecular targets associated with cancer cells—most often proteins expressed on the cell surface. One of the earliest identified targets is the human epidermal growth factor receptor 2 (HER2), a protein from the epidermal growth factor receptor (EGFR) family. HER2 overexpression has been detected in breast cancer cells, as well as in gastric cancer and certain rare subtypes of lung cancer.
In the case of breast cancer, HER2 overexpression is associated with more aggressive tumor growth and a poorer prognosis (an unfavorable prognostic factor), while also enabling targeted therapy using a monoclonal antibody directed against this protein (a favorable predictive factor). Currently, HER2 expression in breast cancer cells is determined using the immunohistochemical (IHC) method and—in uncertain cases—fluorescence in situ hybridization (FISH) on tumor tissue obtained via biopsy or surgery. The IHC method uses a chemical stain to evaluate the reaction result on a scale of 0-3+. A score of 0-1 is considered HER2-negative, while a score of 2+ requires confirmation using FISH (a positive FISH result qualifies the tumor as HER2-positive). A score of 3+ indicates positive HER2 expression. Recently, the concept of HER2-low has been introduced, referring to low HER2 expression that is undetectable using standard methods but still considered a positive predictive factor, meaning it serves as an indication for targeted therapy against this protein. The classification change in HER2 protein expression is illustrated in the figure below [2].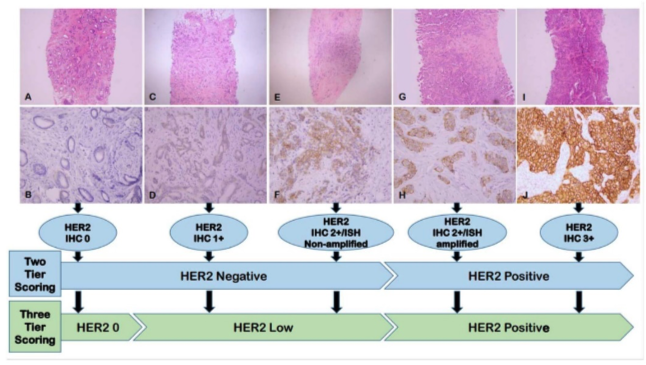
Fig. 1 Current paradigm for assessing HER2 overexpression [2]
The decision made on April 5, 2024, by the US Food and Drug Administration (FDA) to grant accelerated approval for the medicinal product fam-trastuzumab deruxtecan-nxki (Enhertu, Daiichi Sankyo, Inc.) "for adult patients with unresectable or metastatic HER2-positive (IHC3+) solid tumors who have previously received systemic treatment and have no satisfactory alternative therapeutic options" further highlights the need for reliable methods of determining HER2 expression [3].
It should also be emphasized that regardless of changing views on HER2 protein expression in cancer cells, the obtained result is subjective, which naturally entails the risk of obtaining imprecise results. This, in turn, may lead to inadequate/suboptimal treatment for patients with a false-negative result or unnecessary, excessive treatment in the case of a false-positive result.
SDS Optic SA from Lublin has developed the inPROBE® technology platform, designed specifically for detecting protein expression in a way different from the standard approach. First, the detection is performed in vivo, meaning directly within the patient’s body, without the need for tissue sampling. Second, it can be carried out both within the tumor itself and in its immediate surroundings. Third, the result is obtained in real-time, approximately within 30 minutes. And finally, the result is quantitative, meaning it is objective and expressed in numerical values.
The technology platform developed by SDS Optic is based on a combination of fiber optic technology and sensors functionalized with proteins that recognize a specific diagnostic biomarker, such as the HER2 protein. The optical signal generated by the interaction between the protein and the sensor surface is converted, using a mathematical algorithm, into a numerical value representing the protein expression level.
The first protein for which an appropriate inPROBE® probe was developed was HER2. To confirm the feasibility of using inPROBE® in clinical practice and to assess the safety of this new technology, an "Open, multicenter, single-arm clinical study on the safety and efficacy of the inPROBE® diagnostic microprobe for evaluating HER2 receptor expression in a population of women at high risk of breast cancer" (SDS-HER-01-2018) was conducted, with the first part recently completed.
The study included 22 patients, and for statistical analysis, a total of 18 adult patients with a diagnosis of breast cancer and known HER2 expression status, determined using standard methods, i.e., immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH), were included. The study group consisted of 6 patients with HER2-positive and 12 patients with HER2-negative breast cancer according to IHC/FISH results.
The primary endpoint of the study was the preliminary determination of the range of HER2 receptor concentrations measured using the inPROBE® microprobe corresponding to the HER2 receptor status determined by the standard diagnostic method (IHC and FISH tests).
The ranges of HER2 receptor concentrations measured using the inPROBE® microprobe (for which the median, mean, and minimum and maximum values were calculated) were compared with the HER2-positive and HER2-negative status determined by standard methods. A clear numerical trend was observed toward higher values in the HER2-positive group compared to the HER2-negative group, despite the small sample size of the entire study population (which is typical for early-phase studies) and a larger number of patients with HER2-negative tumors. Statistical significance was achieved for the minimum values of the concentration ranges measured using the inPROBE® probe (Wilcoxon rank-sum test; p=0.041). These observations were confirmed in an analysis conducted using linear mixed models for repeated measurements.
Analyzing the obtained results, attention was drawn to the impact of the size of the entire group and individual subgroups. However, the current state of knowledge indicates that the group of HER2-negative patients likely represented a heterogeneous population, which also included patients with the HER2-low characteristic. It can therefore be assumed that the inPROBE® technology developed enabled the detection of a certain biological feature of breast cancer, which was later confirmed in large, international studies [2, 4]. For this reason, in the second part of the study, appropriate modifications will be introduced to the protocol, allowing for a more precise evaluation of IHC staining and the adjustment of HER2 expression markers using the inPROBE® probe to the current classification of HER2 expression.
The secondary endpoint of the study was to compare the correlation of HER2 receptor concentrations detected by one inPROBE® microprobe located in the tumor and another microprobe in the immediate vicinity of the tumor in HER2-positive patients. Although this endpoint was not achieved, likely due to the small number of HER2-positive cancer patients (n=6), a statistically significant correlation was demonstrated between the HER2 receptor concentrations detected using the inPROBE® microprobe in the tumor and in the immediate vicinity of the tumor for the entire study population (both HER2-positive and HER2-negative patients) (p=0.046).
In part I of the clinical study, the safety of the procedure was also assessed. During the diagnostic examination, no defects, damage, malfunctions, or cracks of the inPROBE® microprobe were observed. No safety-related events associated with the device or any adverse effects were reported for the patients participating in the study.
The results of the first part of the clinical study on the measurement of HER2 protein expression in breast cancer justify the initiation of further research projects. These will include other cancers that exhibit HER2 expression, the assessment of protein levels related to viral infections, and the examination of other types of biological material, such as collected tissues or body fluids. The data collected so far suggest that the inPROBE® technology platform is a multipotential, effective, and safe method for the quantitative determination of various proteins in biological material.
It also seems that it may have broader applications not only in medicine, where it can be used for drug development and determining their pharmacokinetics, but also in other areas of science that benefit from the advances in molecular biology and genetics, such as agriculture, veterinary medicine, forensics, and the food and chemical industries.
Literature
1. Iqbal Ni, Iqbal Na. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol Biol Int 2014; 2014: 852748. 2. Zhang H, Peng Y. Current Biological, Pathological and Clinical Landscape of HER2-Low Breast Cancer. Cancers 2023, 15, 126. 3. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-fam-trastuzumab-deruxtecan-nxki-unresectable-or-metastatic-her2 [Dotęp: 07.04.2024] 4. Tarantino P, Hamilton E, Tolaney SM, et al. HER2-Low Breast Cancer: Pathological and Clinical Landscape. J Clin Oncol 2020; 38(17): 1951.
We Care About Your Privacy
SDS Optic S.A. uses cookies to improve and customize users experience on our website. By selecting 'Accept', you consent to the use of all cookies that gather and use information about your interactions with our site to provide personalized content and enhance your digital experience. Please read our Privacy Policy for more information.